Profiling of standard mouse strains
Mice have been widely used as models of human diseases because genetic modification in mice can be easily applied. Many of the mouse strains maintained in our laboratory animal resource bank are genetically modified mice. Inbred mice such as C57BL/6, 129, and FVB have been used, and a variety of model mice have been created, but sometimes you may not get the expected phenotype. It is well known that mice have strain differences and that the phenotype varies greatly depending on the strain used as the genetic background. For example, comparing the total cholesterol in the serum of six strains shows that there are strain differences.
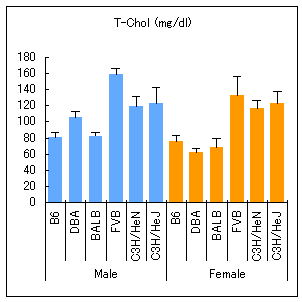
Total chlesterol
B6: C57BL/6JJcl, DBA: DBA/2JJcl, BALB: BALB/cAJcl, FVB: FVB/NJcl, C3H/HeN: C3H/HeNJcl, C3H/HeJ: C3H/HeJJcl
All mice were purchased from CLEA Japan at the age of 8 weeks. For four weeks from 8 to 12 weeks of age, the mice were kept with feed CE-2 (CLEA Japan) at the animal facility of National Institutes of Biomedical Innovation, Health and Nutrition. After 16 hours of fasting, blood was collected from eight males and eight females per strain. Serum biochemical values were measured in a dry-chem 7000V (Fujifilm). Values are means ± SD.
→ Click here for other serum biochemical data.
The graphs below show the results of a comparison between C57BL/6J (B6) and DBA/2J (DBA2), which also shows strain differences in weight change. Body weights of eight males and eight females per strain were measured after regular diet CE-2 or high-fat diet QF had been given for four weeks.
Higher weight gain was observed in mice on high-fat diets. Notably, the strain difference was more evident in females than in males.
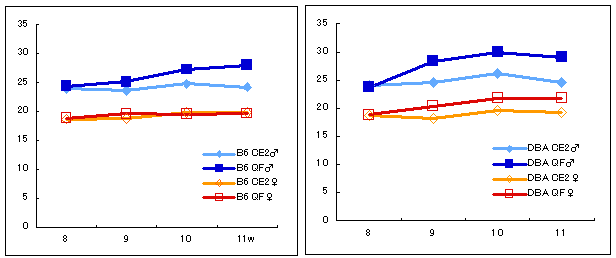
→ Click here for body weight changes in other strains.
In addition, there are strain differences in the amount of genes expressed in each organ. As an example, the results of comparing gene expression of UCP1 in brown fat are shown below. UCP uncouples mitochondrial oxidative phosphorylation and dissipates energy as heat. UCP1 in brown adipocytes is known to decrease in obesity.
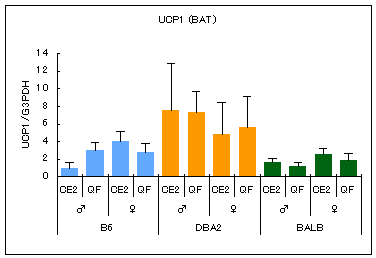
B6: C57BL/6JJcl, DBA: DBA/2JJcl, BALB: BALB/cAJcl
All mice were purchased from CLEA Japan at the age of 8 weeks. For four weeks from 8 to 12 weeks of age, the mice were kept with feed CE-2 (CLEA Japan) at the animal facility of National Institutes of Biomedical Innovation, Health and Nutrition. After 16 hours of fasting, RNA was extracted from brown fat and analyzed by Real Time PCR using TaqMan probe. Values are means ± SD.
→ Click here for other gene expression data (under construction).
By utilizing such strain differences in mouse phenotypes, it is possible to create a variety of disease models that can be applied to the diversity required for disease research, that is, racial differences and individual constitutional differences. In addition, it is very efficient if you can select a mouse strain that has the property of being susceptible to the target disease before creating genetically modified animals. It is known that environmental factors such as diet and breeding method greatly affect the phenotype, so it is also effective to combine genetic background with environmental factors for creating better animal models.
How do you change the genetic background of mice?
The genetic background other than the mutant gene is replaced by at least 12 times of backcrossing in the line to which the mutant gene, etc. are introduced (congenic). It will take more than three years. However, the speed congenic method using microsatellite markers can be completed in 6-7 crosses (about a year and a half). Existing databases are available for genetic polymorphisms among inbred mice.
Please refer to the following database MMDBJ for more information about microsatellite markers.
http://www.shigen.nig.ac.jp/mouse/mmdbj/top.jsp
Nowadays, the number is low, but sometimes hybrid strains (e.g., BDF1) are used to create transgenic mice, and ES cells derived from 129 strains are used to create knockout mice. In such cases, for example, repeat back-crossings with C57BL / 6 mice are necessary to make congenic strains before using it for pathological analysis. The speed-congenic method is also a very effective method. Satellite markers are often used in linkage analyses to identify the causative gene in mouse models with spontaneous mutations.
In addition, satellite markers can be detected more easily by using the multiplex PCR method. As an example, the results of multiplex PCR for D1Mit200, D1Mit253, D1Mit206, and D1Mit217 (see the above database for primers) are shown. As shown in the picture below, you can efficiently obtain the results of multiple markers at the same time.
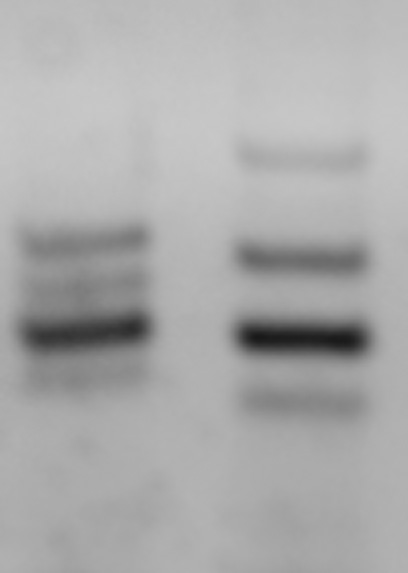
3.5% agarose gel electrophoresis
Left:C57BL/6J, Right:DBA/2J. Bands from top to bottom: D1Mit200, D1Mit217, D1Mit253, D1Mit206.
Why don't you make use of the diversity of mouse strains to create mouse models of human diseases?
